Welcome to RMBase v2.0 !
Latest version of RMBase (RMBase v3.0) is now available at https://rna.sysu.edu.cn/rmbase3/ or http://bioinformaticsscience.cn/rmbase/.
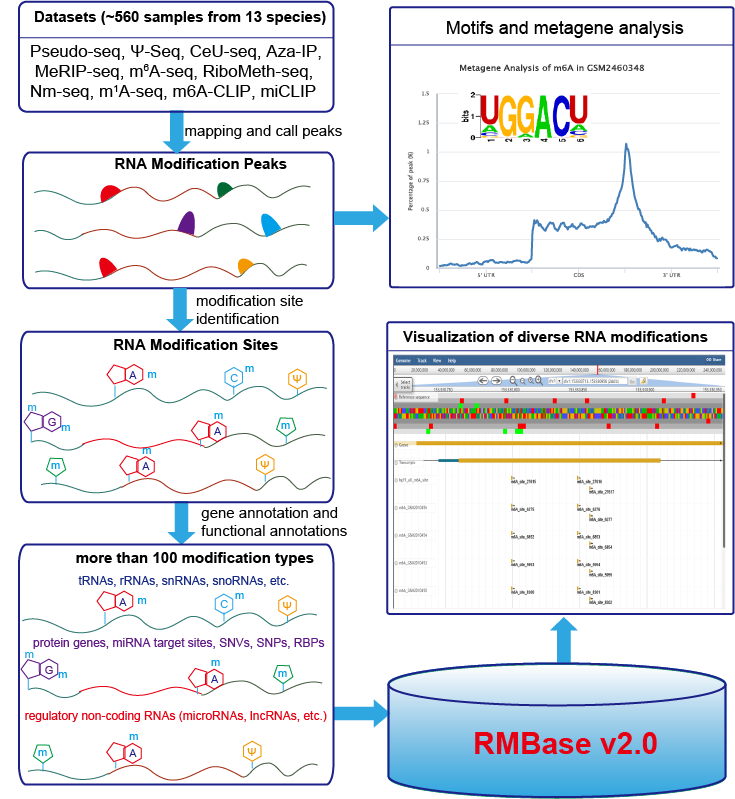
RMBase v2.0 is a comprehensive database to integrate epitranscriptome sequencing data for exploring post-transcriptionally modifications of RNAs, as well as their relationships with microRNA binding events, disease-related SNPs and RNA-binding proteins (RBP). RMBase v2.0 was expanded with 566 datasets and 1 397 244 modification sites from 47 studies among 13 species, which represented about 10 times expansion when compared to the previous release. It contains ~1 373 000 N6-Methyladenosines (m6A), ~5 400 N1-Methyladenosines (m1A), ~9 600 pseudouridine (Ψ) modifications, ~1 000 5-methylcytosine (m5C) modifications, ~5 100 2′-O-methylations (2′-O-Me), and ~2 800 modifications of 100 other types.
We built two new modules called “Motif” and “modRBP”, respectively. “Motif” provides both de novo identified position weight matrices (PWMs) and visualized logos of modification motifs. And “modRBP” analyzes the relationships of RNA modifications and RNA-binding Protein (RBP). We also constructed a new web interface, "modSNV", to present the disease link between RNA modification sites and SNVs sites. In addition, we developed a noval web-based tool named "modMetagene" for plotting metagenes of RNA modifications along a transcript model.
RMBase v2.0: Deciphering the Map of RNA Modifications from Epitranscriptome Sequencing Data. Jia-Jia Xuan, Wen-Ju Sun, Ke-Ren Zhou, Shun Liu, Peng-Hui Lin, Ling-Ling Zheng, Liang-Hu Qu*, Jian-Hua Yang*.
RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Wen-Ju Sun, Jun-Hao Li, Shun Liu, Jie Wu, Liang-Hu Qu*, Jian-Hua Yang*. Nucleic Acids Res. 2016 Jan 4;44(D1):D259-65. Epub 2015 Oct 12.
Release notes
Release version 2.0.0 2017-06-01
- Species: 13
- Sample: 566
- modification sites: 1,397,244
- Technologies: Pseudo-seq, Ψ-seq, CeU-seq, Aza-IP, MeRIP-seq, m6A-seq, m1A-seq, miCLIP, m6A-CLIP, RiboMeth-seq
- Modification Types: over 100
- located ~5,400 m1A (1-methyladenosine) modification sites by de novo analyzing from 4 species.
- de novo identified ~1,373,000 m6A (N6-Methyladenosines) modification sites among 12 species.
- Expanded to ~5,100 2′-O-Me (2′-O-methylations) modification sites.
- increased to ~9,600 Ψ (pseudouridine) modification sites.
- built a new module, Motif, presenting PWMs with motif logos and metagene.
- developed "modMetagene", a new tool for metagene analysis on user uploaded data.
- constructed "modRBP", which analyzed the relationships of RNA modifications and RNA-binding Proteins (RBPs).
- analyzed relationships of RNA modifications and SNVs and found that at least 10 RNA modification types were potentially correlated with dozens of diseases or cancers.
Modification Site Statistics in 13 Species
Rattus
Pig
Zebrafish
A.thaliana
S.cerevisiae
Fly
P.aeruginosa
E.coli
S.pombe
RNA Modification Types Statistics
| m1A | m5C | m6A | 2'-O-Me | pseudoU | other |
|---|---|---|---|---|---|
| 5411 | 988 | 1373355 | 5096 | 9570 | 2824 |
| 0.387% | 0.071% | 98.290% | 0.365% | 0.685% | 0.202% |
Contact us:
RMBase is under constant maintenance and improvement. Any questions about the usage, comments or suggestions are appreciated.
Please contact us: yangjh7@mail.sysu.edu.cn
Jian-Hua Yang
RNA Information Center, State Key Laboratory for Biocontrol, Sun Yat-sen University, Guangzhou 510275, P. R. China
For better browse experience, Chrome is strongly recommanded. Firefox 37, Opera 28, Safari 5 and Internet Explorer 9-11 are also supported with some display bugs and less compatibility.