Enzymes 
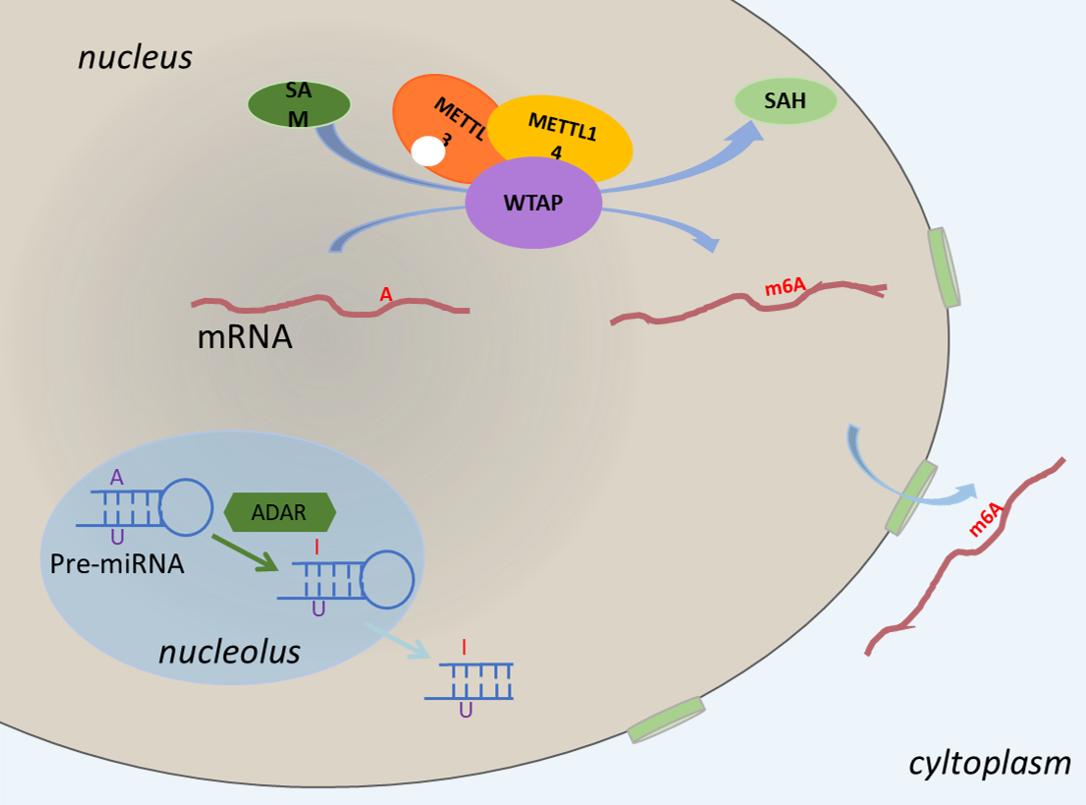
RNA modification is a dynamic process which occurs under the function of some enzymes whose action is similar to that of "writers". RNA modifications are placed under 3 groups, namely methylation that contains ribosomal methylation, base methylation and mixed methylation, pseudouridine and hypoxanthine respectively.
The overview of RNA modification sites guided by enzymes as follows:
| ADAR | FBL | METTL14 | METTL3 | NOP56 | NOP58 | NSUN2 | PUS1 | RBM15B | RBM15 | WTAP |
|---|---|---|---|---|---|---|---|---|---|---|
| RNA editing | Nm | m6A | m6A | Nm | Nm | m5C | Pseudo | m6A | m6A | m6A |
| 1462 | 1894 | 1504 | 1726 | 1306 | 1863 | 29 | 25 | 8404 | 13565 | 573 |
snoRNAs 
C/D box snoRNAs
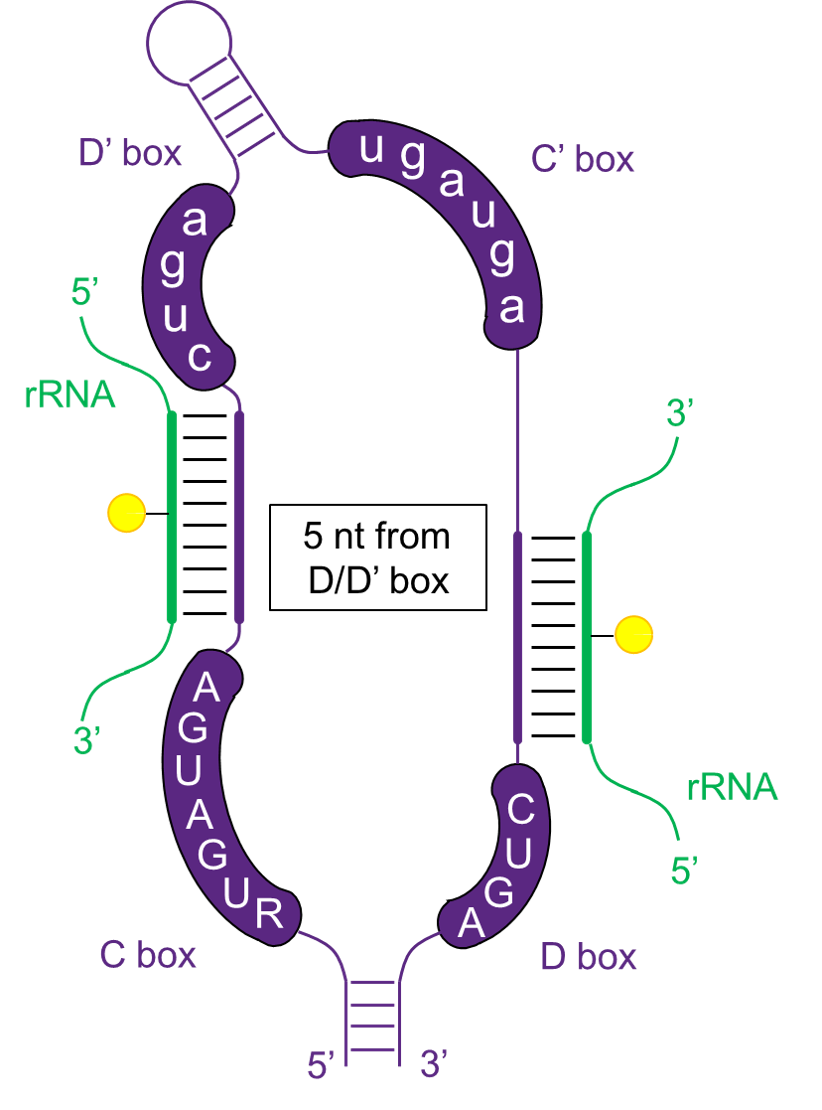
The C/D box snoRNAs carry two conserved boxes, C box with 'RUGAUGA' in 5' end and D box with 'CUGA' in 3' end. While C'/D' box, the copies of C/D box are usually inside snoRNAs.
C/D box snoRNA binds to proteins to form complexes, snoRNPs, that catalyze RNA modifications. These proteins include FBL, NOP56, NOP5/NOP58 and NHPX. C/D box snoRNAs guide 2'-O-ribose methylation of target RNAs by upstream components of D/D' box.
H/ACA box snoRNAs
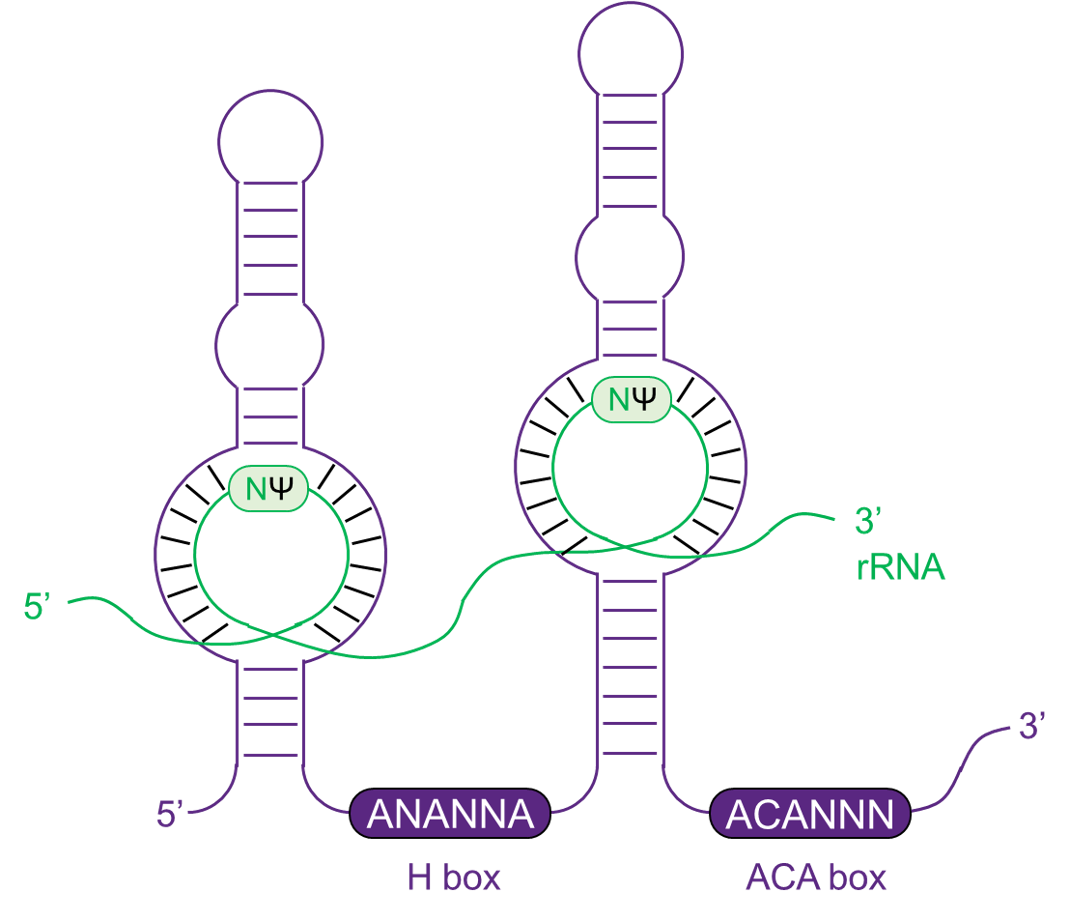
The H/ACA box snoRNAs contain two hairpins and two single-stranded regions. The single-stranded regions contain H box with 'ANANNA' internally and ACA box with 'ACA' in 3' end. While the bulges of hairpins bind to target RNA and guide pseudouridine modification of specific loci.
H/ACA box snoRNA binds to four proteins, including dyskerin, GAR1, NHP2 and NOP10 to guide pseudouridine methylation of target RNAs.
Cajal body-specific scaRNAs
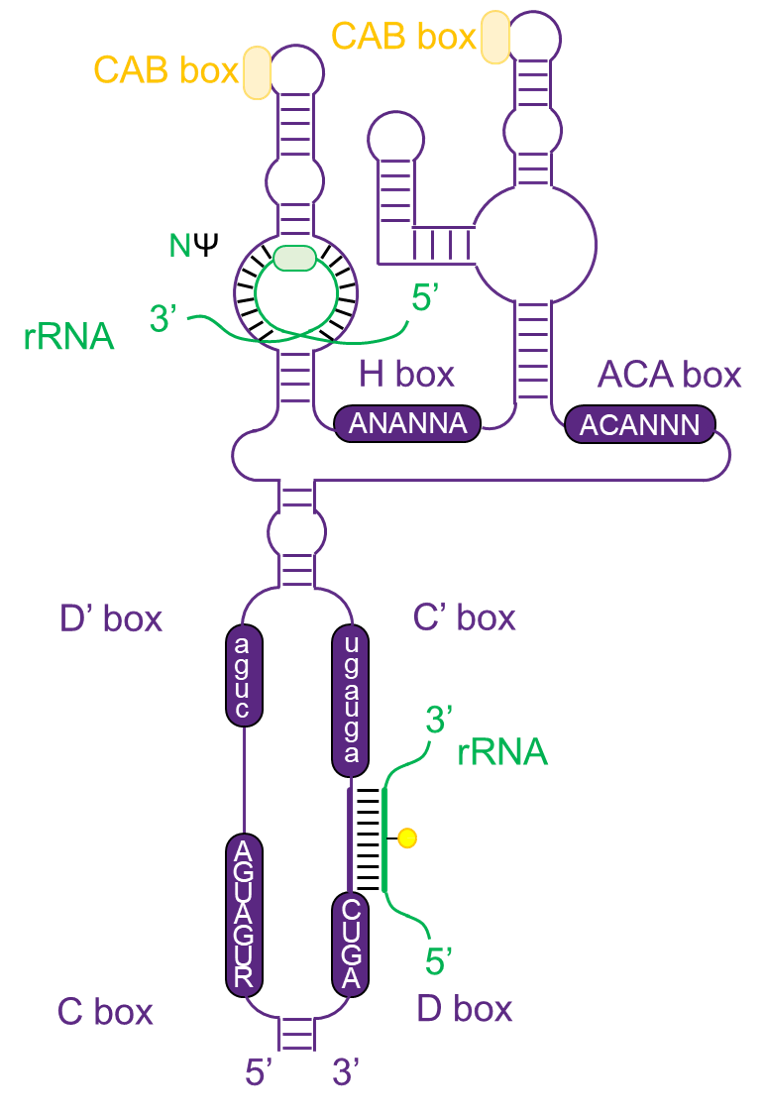
The Cajal body-specific scaRNAs have more complex chemical structure that offen contain both C/D box and H/ACA box. They can guide modifications of snRNA, like U1, U2, U4 and U5 that transcribed by polymerase II.
The overview of RNA modification sites guided by snoRNAs as follows:
| Human | Mouse | Yeast | Fly | C.elegans | Zebrafish | A.thaliana | |
|---|---|---|---|---|---|---|---|
| Pseudo | 1574 | 659 | 98 | 342 | 144 | 5 | 28 |
| Am | 631 | 127 | 42 | 174 | 40 | 9 | 26 |
| Cm | 390 | 66 | 22 | 76 | 55 | 0 | 26 |
| Gm | 826 | 85 | 35 | 143 | 28 | 18 | 17 |
| Um | 185 | 37 | 23 | 48 | 25 | 11 | 33 |