Welcome to RMBase v3.0!
If slow, you can access it Here.
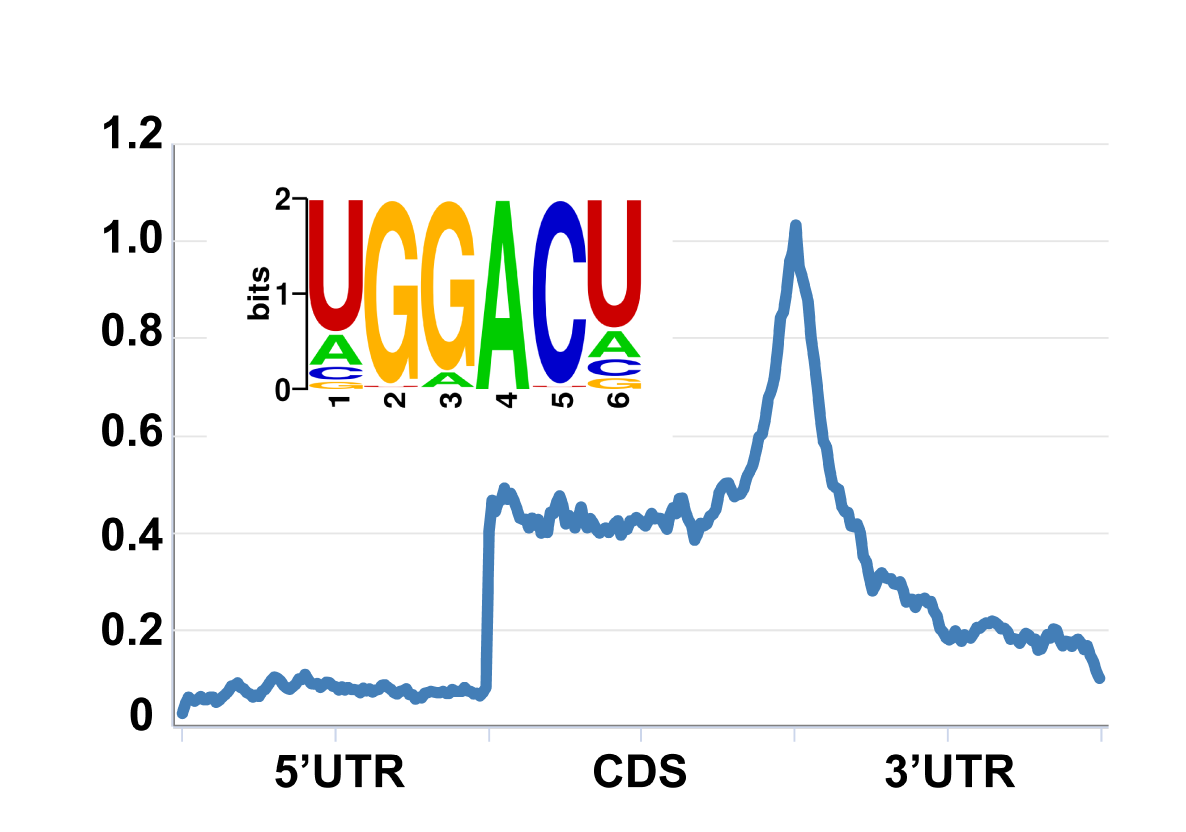
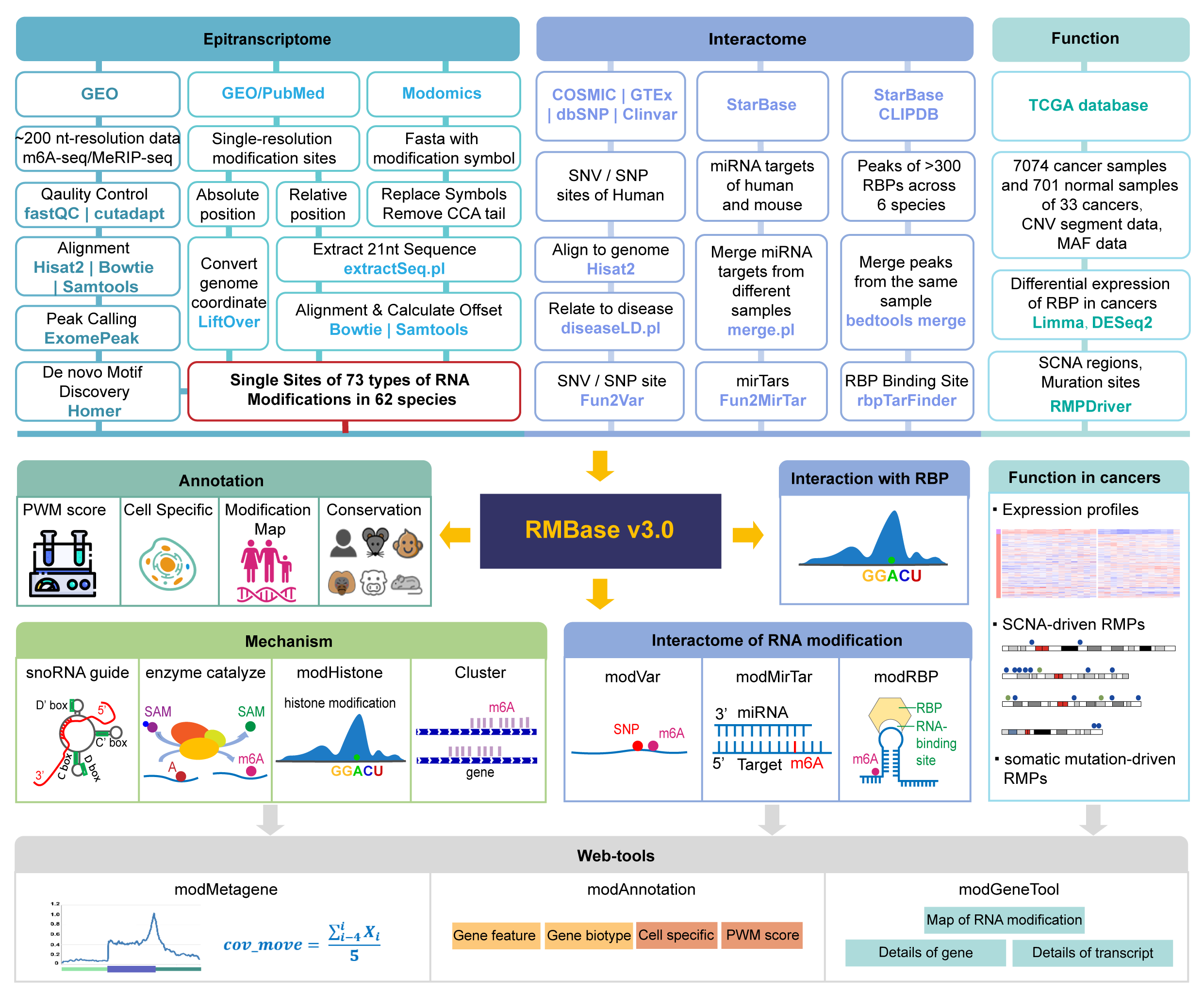
RMBase v3.0 is an upgraded version of RMBase that mainly focuses on the mechanism and function of diverse RNA modifications. RMBase v3.0 is a comprehensive and convenient platform for efficient studying RNA modifications from large amounts of epitranscriptome high-throughput sequencing data. It provides multiple interfaces and web-based tools to integrate 73 types of RNA modification among 62 species, uncover the relationships between RNA modifications with a series of interacting factors, and reveal the distribution patterns, biochemical mechanisms, evolutionary conservation of RNA modifications and their biological roles in human diseases.
Finally, RMBase v3.0 provides three powerful web-based tools including modAnnotation, modMetagene and modGeneTool for custom analysis of RNA modification data.
The database idetified 1,074,100 RNA modification sites among 73 type of RNA modifications in 62 .
RMBase v3.0 Release Note (Oct 2023):
- Species: 9 clades, 62 species.
- Samples: 828 wild type datasets.
- RNA Modification: 73 types.
- Tools for RNA modifications: 3 tools.
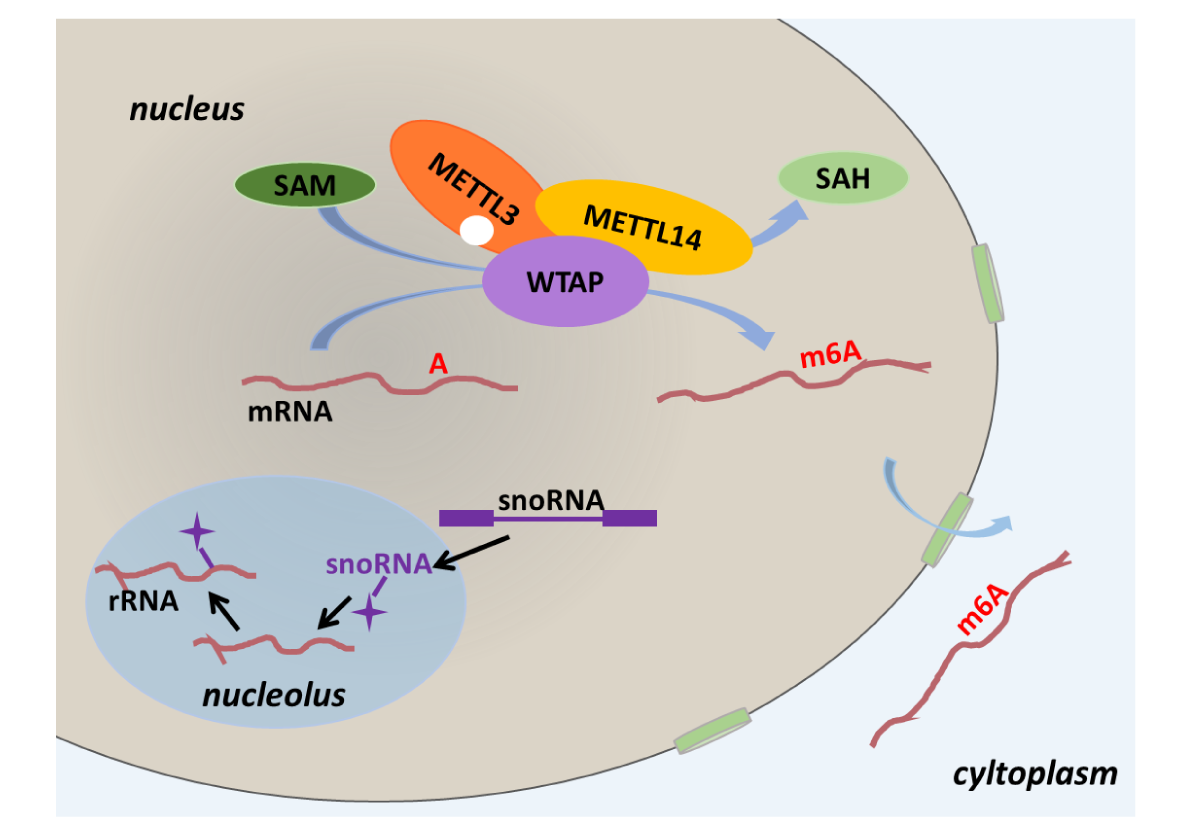
- Explore the pseudo sites guided by known and orphan snoRNAs. These pseudo sites occur on mRNA, rRNA, snRNA, tRNA, lncRNA and repeatMasker.
- Detect the RNA modification clusters in genome wide.
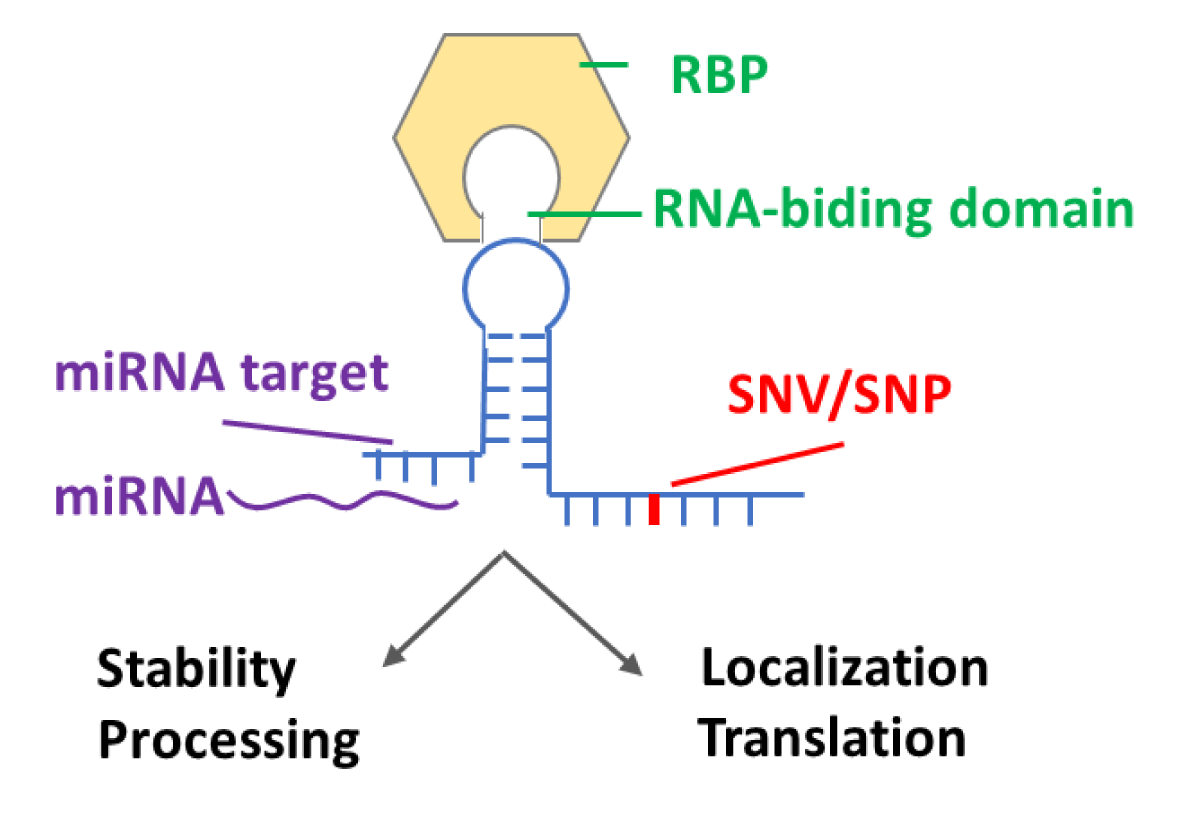
- Decipher the associated functions of RNA modifications with RBP, miRNA target, SNP, and SNV.
- Identify novel m6A readers.
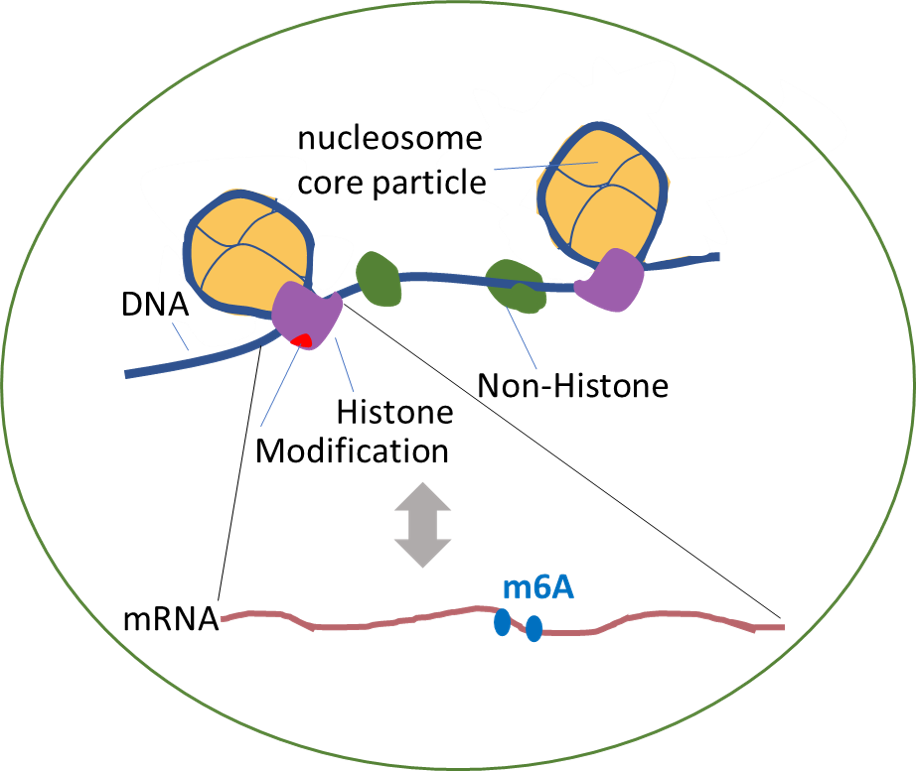
- Explore the potential associations (co-localization) of histone modifications on RNA modifications.
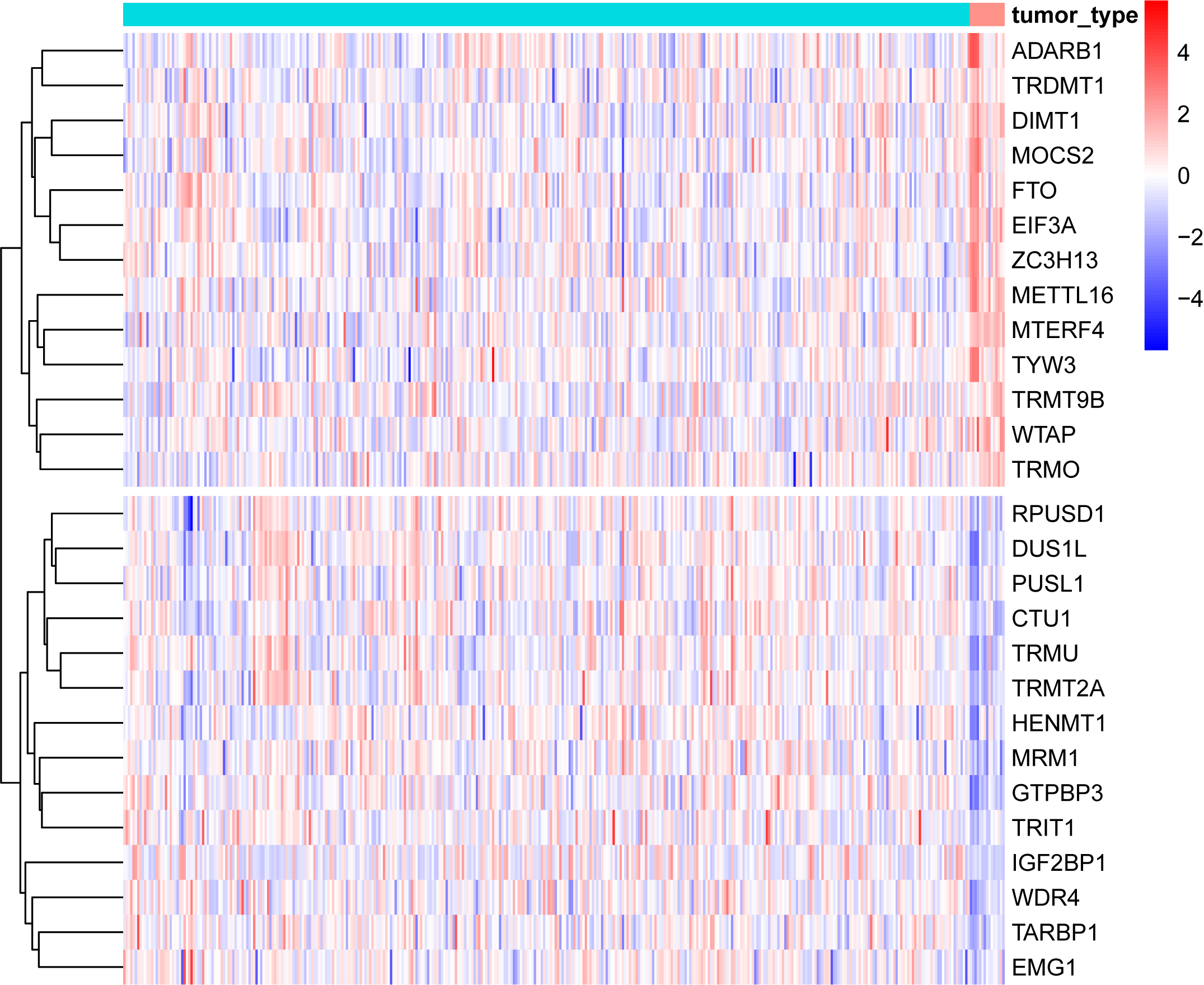
- Investigate the biological functions of RNA modifications in diverse cancers.
How to cite:
- RMBase v3.0: Decode the Landscape, Mechanisms and Functions of RNA Modifications.
- Xuan JJ, et al. RMBase v2.0: Deciphering the Map of RNA Modifications from Epitranscriptome Sequencing Data. Nucleic Acids Res. 2018 Jan 4;46(D1):D327-34.
- Sun WJ, et al. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2016 Jan 4;44(D1):D259-65.