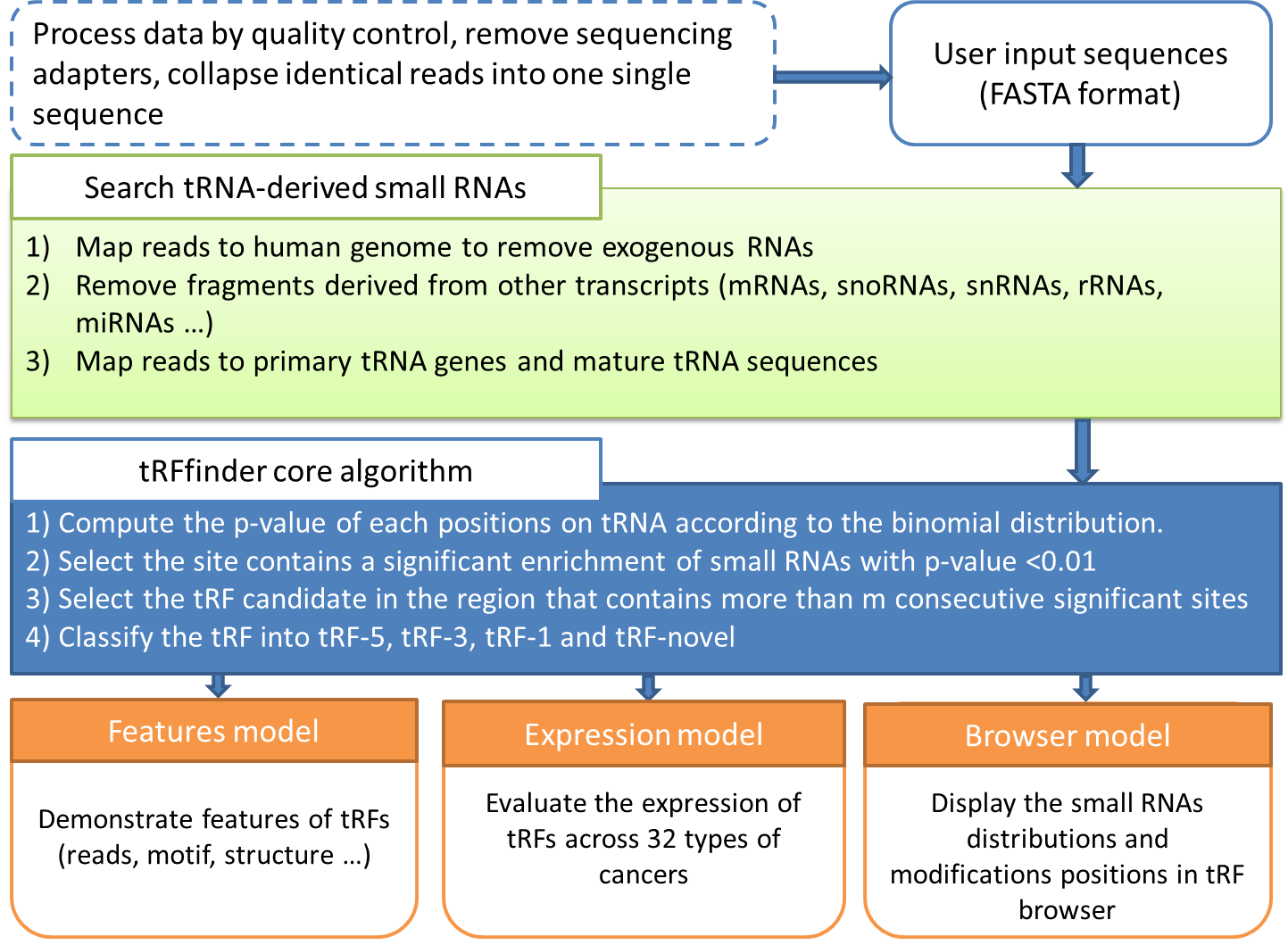
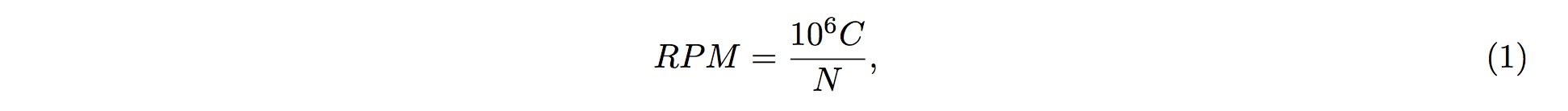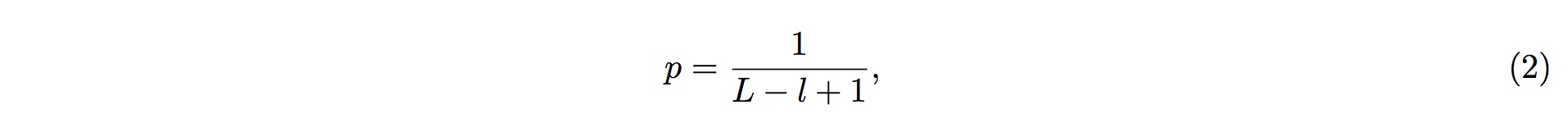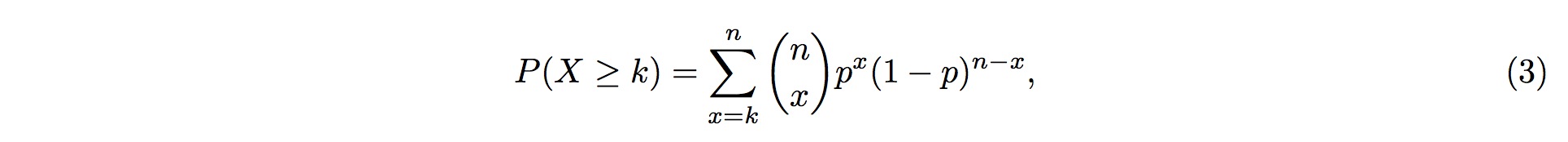To perform the screening, it is first assumed that fragmented RNAs are randomly distributed along the entire pre-tRNA; that is, the RNAs are uniformly distributed across the entire length of pre-tRNA. According to this assumption, we could conclude that, of the entire length of tRNA, the probability of one small-RNA fragment mapped onto one particular position in the tRNA is
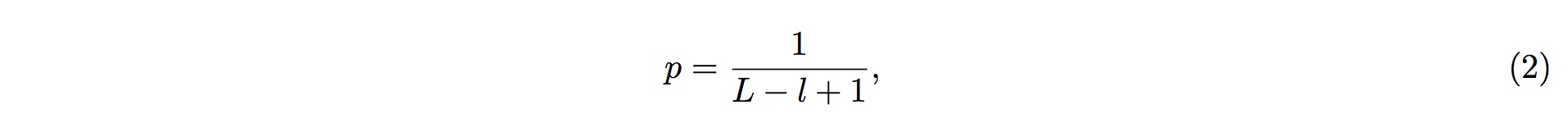
where L is the length (in nucleotide, nt) of the tRNA, and l is the length (in nucleotide, nt) of the small RNA fragment being mapped onto the tRNA.
Therefore, the probability of more than k (inclusive) small RNA fragments mapped onto the same position in the tRNA follows the Binomial distribution, and the probability for this event is
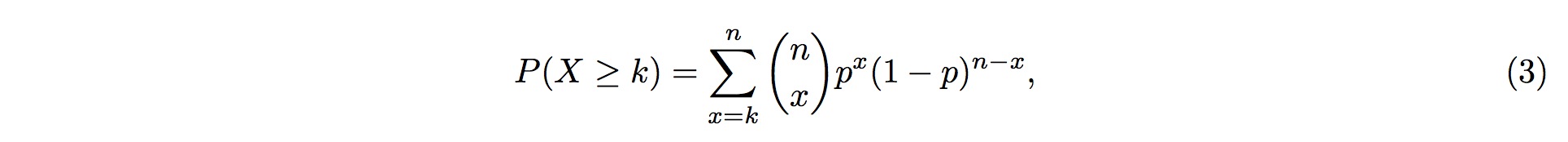
where k is the observed counts of small RNA fragments mapped onto that particular position in the tRNA, and n is the total number of fragments mapped onto the entire tRNA.
If there are more than k (inclusive) small RNA fragments mapped onto one particular position in the tRNA, but the probability of this event occurring by chance (Eq. 2) is less than 1% (referred to the p-value, and can be adjusted to your satisfaction), then we could conclude that the assumption above is false (with 99% confidence, by default); i.e., this event does not occur by chance.
Note, however, that generally tRFs is of more than 16 nt in length. To take this fact into consideration, we should ensure that the tRFs candidate matches with tRNA sequence for consecutive more than 16 nt. (Mismatches, indels are allowed.) This, in turn, corresponds to the requirements that there are at least 16 nt bases in consecutive position in tRNA (which matches with the candidate tRFs sequences) should have a p-values less than 1%. (Figure 2)
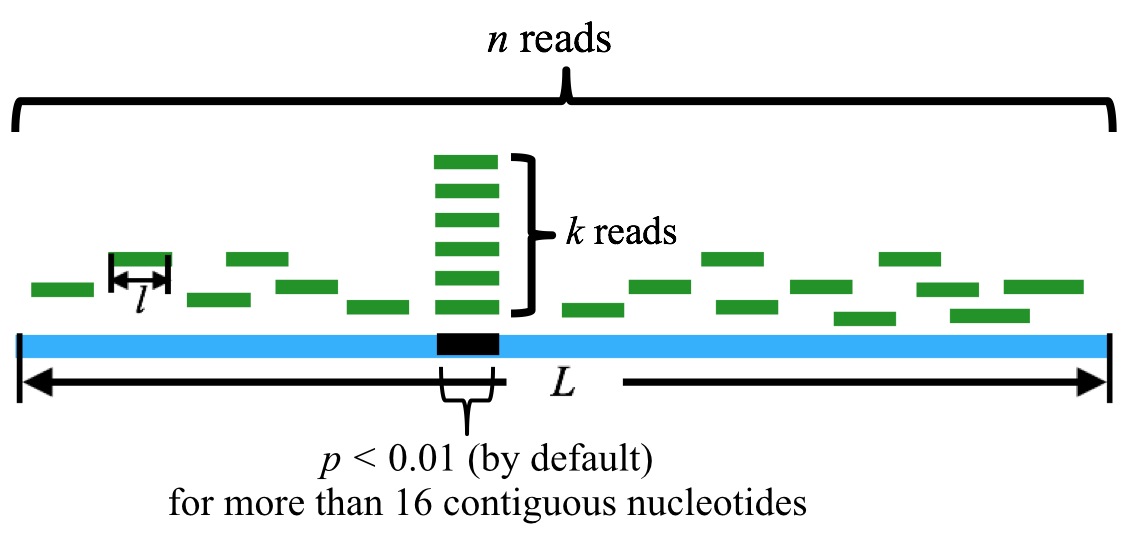
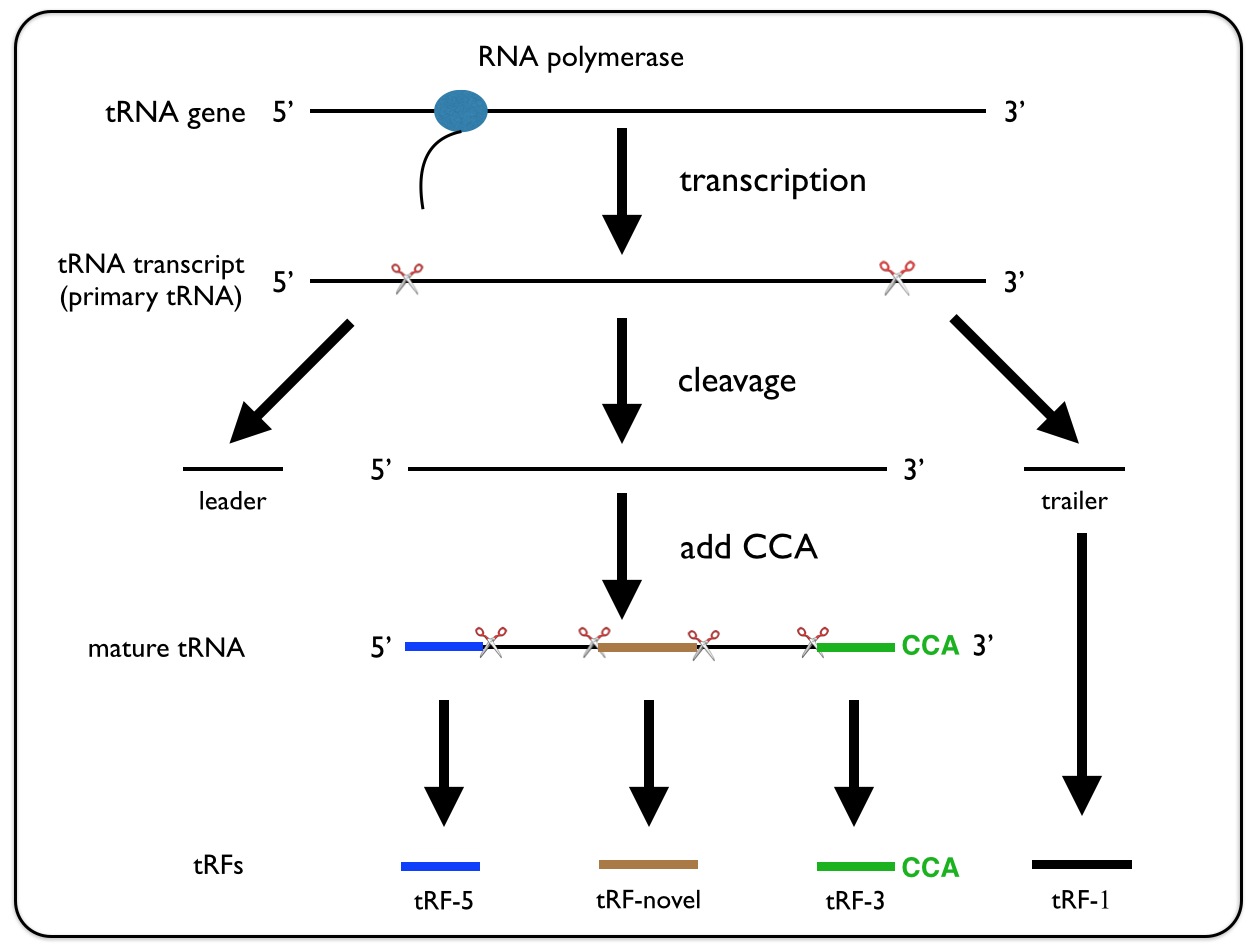
Figure 2. Schematic demonstration of how the core algorithm works.
n is the sum of the reads mapped to the tRNA, k is the sum of the reads that are mapped to the particular position in the tRNA. l is the length of the reads, and L is the length of the tRNA. By default, a tRF candidate corresponds to more than 16 contiguous nucleotides with p-value(s) less than 0.01.