
Welcome to ChIPBase!
News: ChIPBase v2.0 has been updated to v3.0 !
Jumping to ChIPBase v3.0 page to see more function and modules.
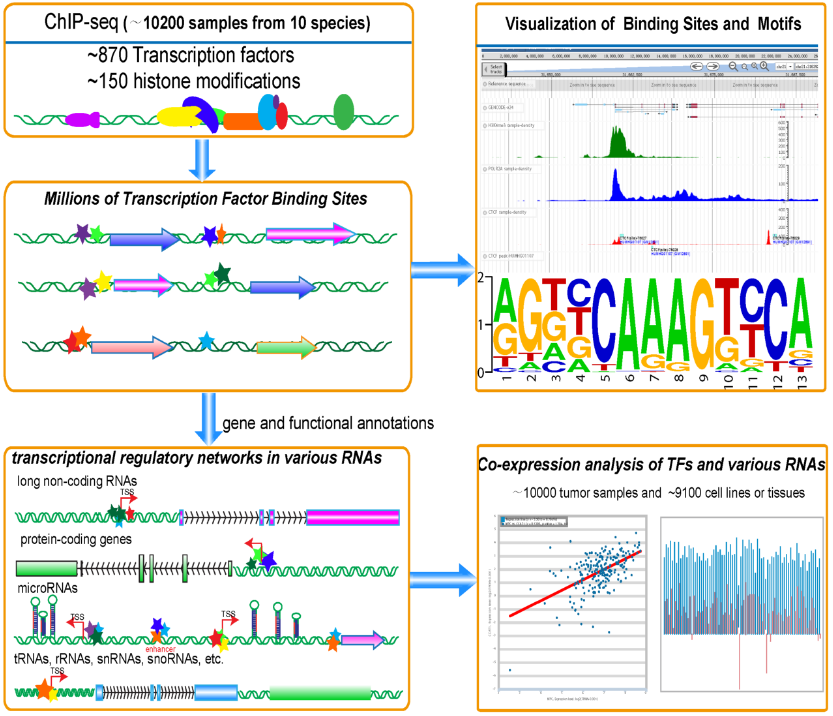
ChIPBase v2.0: decoding transcriptional regulatory networks of non-coding RNAs and protein-coding genes from ChIP-seq data. Zhou KR, Liu S, Sun WJ, Zheng LL, Zhou H, Yang JH*, Qu LH*. Nucleic Acids Res. 2017 Jan 04;45(D1):D43-D50.
ChIPBase: A database for decoding the transcriptional regulation of long non-coding RNA and microRNA genes from ChIP-Seq data. Yang JH, Li JH, Jiang S, Zhou H and Qu LH*. Nucleic Acids Res. 2013 Jan;41:D177-87.Epub 2012 Nov 17
Release notes
Updated gene annotation sets of human and mouse.
- Human: GENCODE v25
- Mouse: GENCODE M10
Release version 2.1.0 (2016-06-01)
Finished data import and website development.
- ChIP-seq data: ~10,000
- RNA-seq data: ~20,000
Sample Statistics in 2 Species
For better browse experience, Chrome is strongly recommended. If you encounter some errors, please clear your browser cache and try again. Firefox 46, Opera 37, Safari 5 and Internet Explorer 9-11 are also supported with some display bugs and less compatibility.