Single cell RNA sequencing (scRNA-Seq) technology has revealed significant differences in gene expression levels between different cell groups. However, most of the studies focus on the analysis of mRNAs, ignore long non-coding RNAs (lncRNAs), which have been shown to be more abundant and have significant cell-specificity. In this study, we developed ColorCells, a platform for comparative analysis of long non-coding RNAs (lncRNAs) and mRNAs expression, classification and functions in single-cell RNA-Seq data. We apply ColorCells to analyze 167913 publicly available scRNA-Seq experiments from 5 species. Integrative annotation of lncRNAs reveals large numbers of cell-specific lncRNAs and their properties. We provides a serious of novel tools and friendly visual interface in ColorCells, including apply PCA and t-SNE algorithm to display cell clusters in 2D and 3D space, develop a tissue map tool to show various tissues and cell types in humans and mouse, establish a statistical test method for hypergeometric distribution to automatically assigncell type labels to cell clusters, estimate cell-cell similarity based on SNN and pearson correlation analysis, built protein-lncRNA co-expression networks to predict lncRNAs function from scRNA-Seq data. Our study emphasizes the need to uncover lncRNAs that occur in all types of cells, and we wish ColorCells to be a good resource for revealing the features, expression and functions of lncRNAs in single cells.
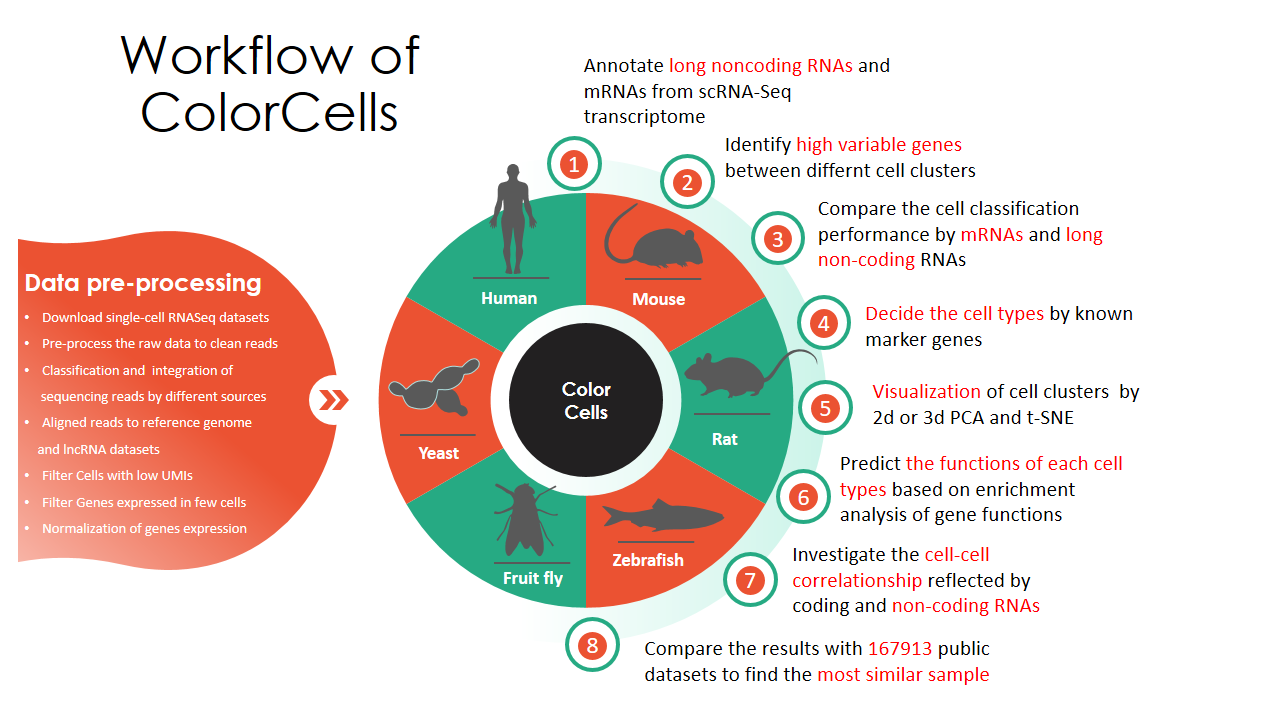
Novel tools in ColorCells:
- Classify single-cells into differnt clusters using lncRNAs and mRNAs
- Automatically assign cell type labels to cell clusters based on statistical test method
- Search for the most similar sample from 167913 public scRNA-Seq datasets by lnRNAs and mRNAs profiles
- Browse public scRNA-Seq studies from human and mouse by tissue map
Special Features:
- Dimensional reduction and visualisation of cell types with PCA and t-SNE
- Cell-cell similarities estimation by shared nearest neighbor (SNN) and pearson correlation
- lncRNAs and mRNAs marker gene expression profiles in each cell cluster
- Enrichment analysis of marker gene functions in each cell cluster