Overview
dreamBase 2.0: DNA regulation, RNA expression and protein translation of pseudogenes in human and mouse.
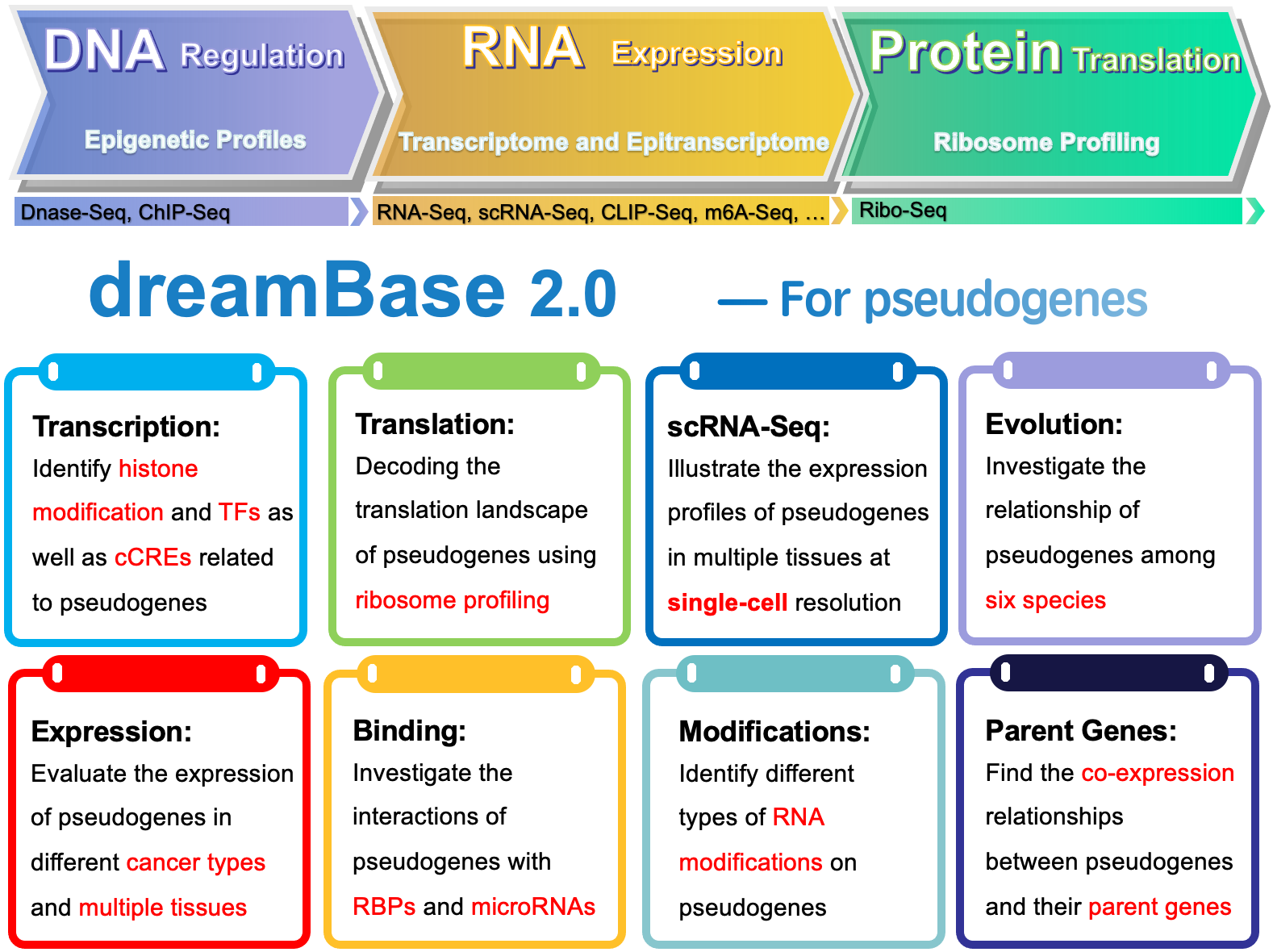
With ~5500 ChIP-seq and DNase-seq data, dreamBase 1.0 has provided genome-wide distribution patterns of Transcription Factors, Pol II, Histone Modifications and DnaseI Hypersensitivity sites around the transcription start sites of pseudognenes.
Based on ~9165 epigenetic profiles data, dreamBase 2.0 demonstrated the chromosomal status and cis-regulatory elements (cCRE) around pseudogenes as well as the binding sites of trans-regulatory elements. By analyzing ~18196 bulk RNA-seq and 933704 single-cell RNA-seq data, dreamBase 2.0 illustrated the expression profile of pseudogenes in different tissues, cell types and diseases. Based on the ribosomal profile data, dreamBase 2.0 analyzed the enrichment of enrichment of the ribosome profilings on pseudogenes.
We developed a tool, co-pseudo, to explore the co-expression relationships between pseudogenes and their parent genes. Through combining AGO-bound CLIP-seq data contained microRNA binding sites with pseudogenes, we showed complex post-transcriptional regulation networks of microRNAs and pseudogenes. We built the ceRNA network to illustrate the crosstalk between pseudogenes and their parent genes through competitive binding of microRNAs. In addition, we also presented the transcriptome-wide protein-RNA interactions between RNA binding proteins (RBPs) and pseudogenes with 568 CLIP-seq data. Cooperated with RNA modification sequencing data, we mapped 11377 RNA modifications sites onto 2346 pseudogenes, including N6-Methyladenosines (m6A), pseudouridine (Ψ) modification and 2’-O-methylations (2’-O-Me) modification sites.
Based on the above information provided by dreamBase 2.0, researchers can unearth truly functional pseudogenes in the genome and get deeper insights into the transcriptional regulation, expression, evolution, functions and mechanisms of these pseudogenes and their roles in multiple biological process and diseases.
dreamBase: DNA modification, RNA regulation and protein binding of expressed pseudogenes in human health and disease. Zheng LL†*, Zhou KR†, Liu S, Zhang DY, Wang ZL, Chen ZR, Yang JH*, Qu LH*. Nucleic Acids Res. 2018 Jan 4;46(D1):D85-D91.