Welcome to ENCORI Pan-Cancer Analysis Platform
Description of The Platform
ENCORI Pan-Cancer Analysis Platform is designed for decoding Pan-Cancer Networks of lncRNAs, miRNAs, pseudogenes, snoRNAs, RNA-binding proteins (RBPs) and all protein-coding genes by analysing their expression profiles across 32 cancer types ( ~10,000 RNA-seq and ~9,900 miRNA-seq samples) integrated from TCGA project.
Our Platform provided the following Cancer-related Analysis Services:
- ENCORI explored the Pan-Cancer network of miRNA-lncRNA, miRNA-pseudogene, miRNA-sncRNA and miRNA-mRNA interactions, which were supported by hundreds of Ago CLIP-Seq experiments.
- ENCORI built the Pan-Cancer ceRNA networks from miRNA-target interactions which were validated by Ago CLIP-seq experiments.
- ENCORI provided the Pan-Cancer maps of CLIP-seq experimentally verified RBP-lncRNA, RBP-pseudogene and RBP-mRNA interactions.
- ENCORI provided a Pan-Cancer analysis platform to freely explore the co-expression networks of 2 candidate genes (miRNA-RNA/RNA-RNA) in 32 types of cancers.
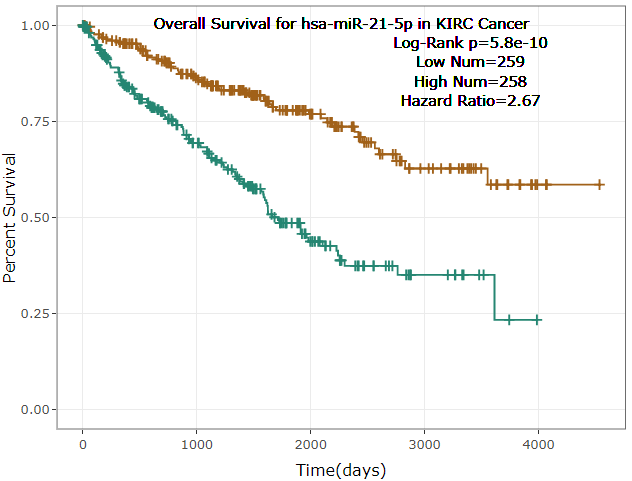
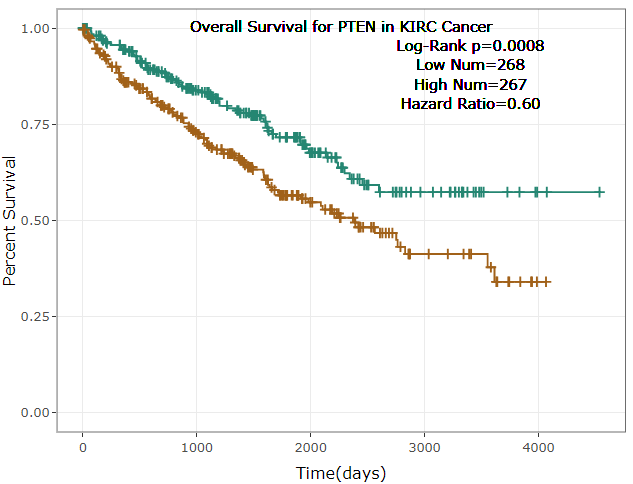
- ENCORI provided survival and differential expression analysis platform for miRNAs and genes(lncRNAs, mRNAs, ncRNAs, pseudogenes, etc.).
- ENCORI provided Scatter Plot, Box Plot and Survival Plot to visualize the above-mentioned pan-cancer maps in 32 types of cancers.
Description:
Here, we provided a query entrance for performing Pan-Cancer analysis on 2 candidate genes (such as miRNA-lncRNA, miRNA-mRNA and RBP-lncRNA) in 32 types of cancers with 10,882 RNA-seq and 10,546 miRNA-seq samples.
The expression data of genes in cancers were downloaded from TCGA project via Genomic Data Commons Data Portal.
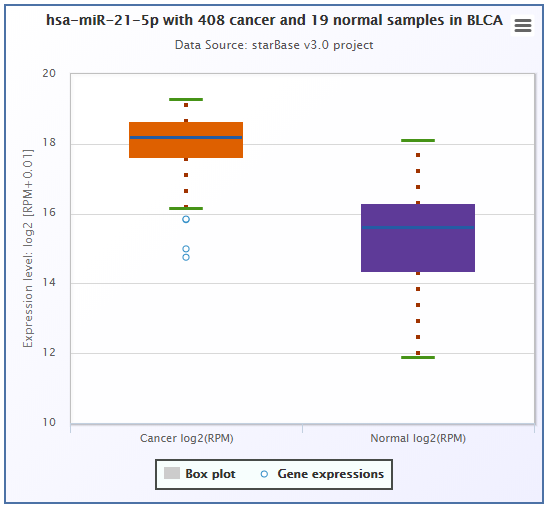
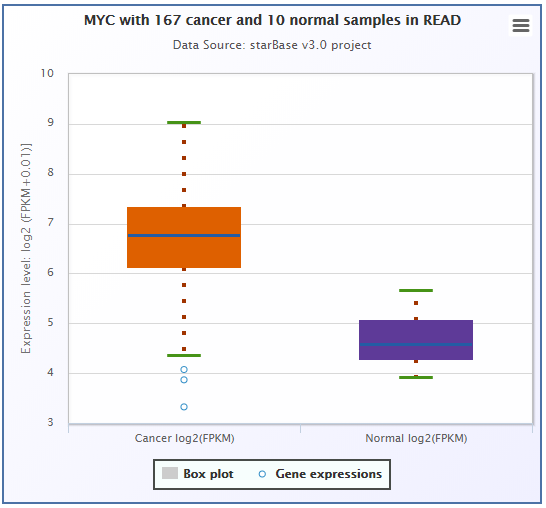
The expression values of genes from RNA-seq data were scaled with log2(FPKM + 0.01), while the ones from miRNA-seq data were scaled with log2(RPM + 0.01).